Biolearn and the Future of Biomarkers¶
The omics revolution is transforming biology by providing a wealth of data that has the potential to transform health care. We see a future where a single blood test could offer comprehensive insights into one’s health and surrogate endpoints for the development of age reversing therapeutics. However, the path to this future is obstructed by challenges such as data access and the complexity of human biology. Biolearn is addressing these issues by developing modern tools for accessing data and extracting insights. Recognizing the magnitude of this challenge, we’ve adopted an open-source and open-science approach. We invite you to explore our roadmap and learn how you can contribute to this mission.
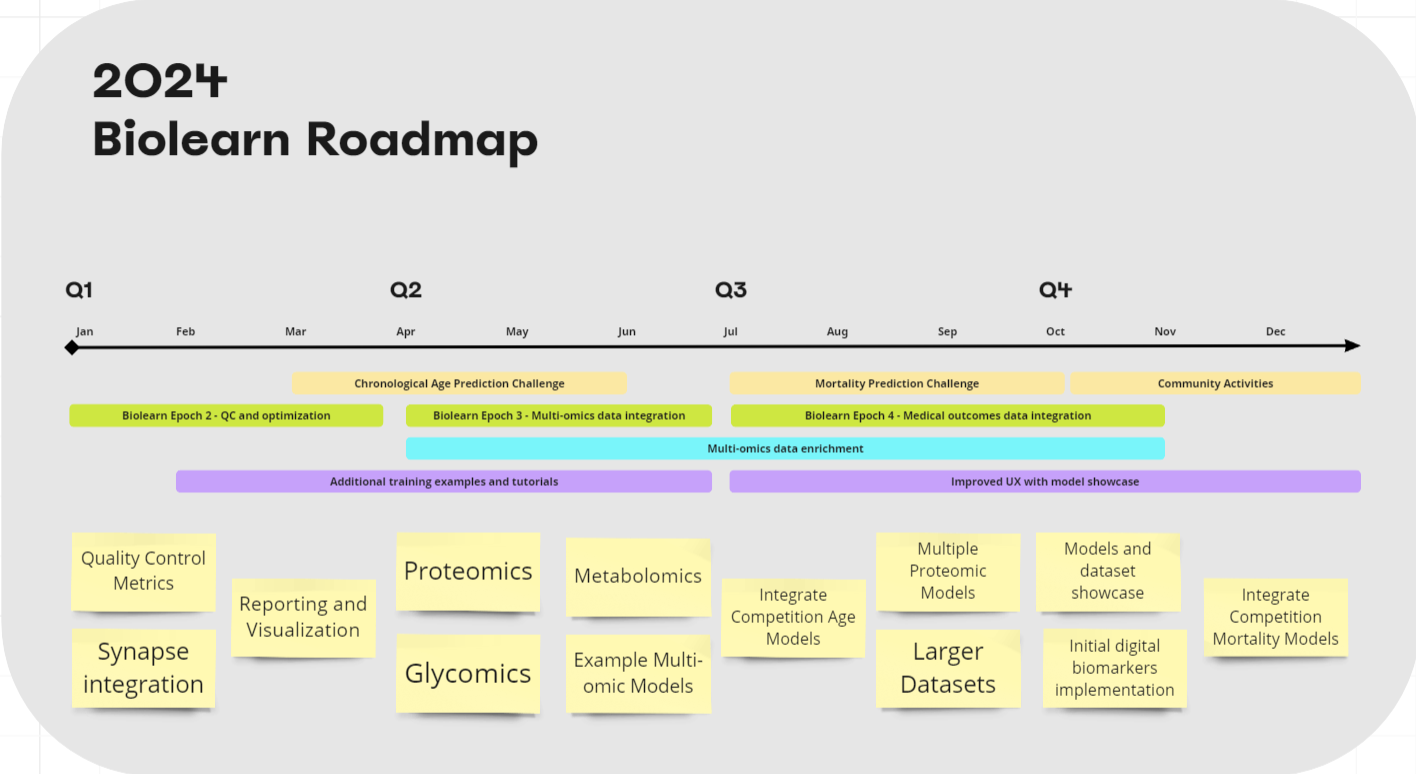
Currently, Biolearn supports primarily epigenetic methylation models with 25 models and easy to load data sources from GEO comprising collectively thousands of samples. We have big ambitions for Biolearn but we need help to achieve them. How can you help?
You can code.
If you can write code we need your help building the library. We have both large and small tasks available for varying skill levels, knowledge of biology and time availability.
Some items needing work right now:
Improved Caching - Caching needs to be better configurable for different environments
Data loading and parsing optimizations - Currently loading large files can take minutes. We can speed this up ~10x.
Integrating blood biomarkers into unified data format - Currently Geo data and blood biomarkers use separate data structures. We want to integrate these in preparation of supporting multi-omics.
Adding GEO datasets - We know of many more GEO datasets that have methylation and age data. This process is partially automated but requires a human to review.
Implement Genetic Disease Models - Currently we have 1 but there are more genetic diseases that can be identified using epigenetic data
You have built a model.
If you have built a model for aging or human health and are interested in sharing it by including it in biolearn please reach out to us. If your model is proprietary but you would be open to sharing access with researchers via API we are interested in talking with you also.
You have access to data.
If you know of useful public omics datasets that are not in biolearn we would like to hear about it. If you have access to data that is currently private but you think could be made more public in some way please reach out to us.
You would like to fund our work.
If you are interested in supporting our mission financially please reach out to us directly. Money will be used to support maintenance and development of Biolearn, provide prize pots for further challenges and support the generation of more expansive open multi-omics datasets.
Join Us¶
If you would like to contriubte join our Discord server and reach out to any of our Maintainers.