Examples¶
Warning
If you want to run the examples, make sure you execute them in a directory where you have write permissions, or you copy the examples into such a directory. If you install biolean manually, make sure you have followed the instructions.
Omics Biomarker Examples¶
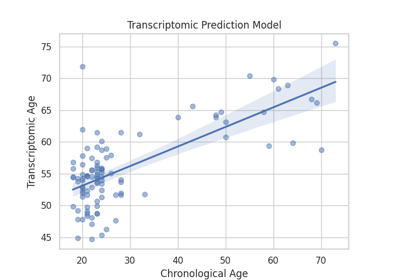
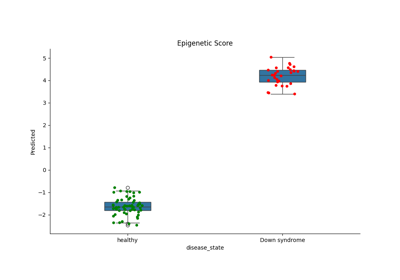
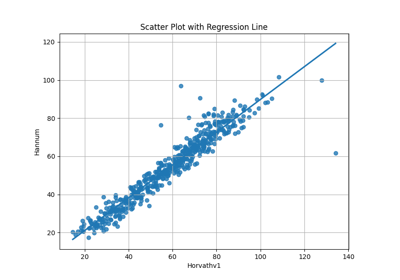
Plots results of running omics based biomarker models functions. Includes methylation and transcriptomic data.
Composite Biomarkers Examples¶
Plot survival curves based on clocks using data from standard blood tests
Challenge Submission Examples¶
Generate submissions for the biomarkers of aging challenge
Building a competition submission using an existing model
Deconvolution Examples¶
Plots results of running deconvolution on dataset with known cell proportions
Additional Visualization Examples¶
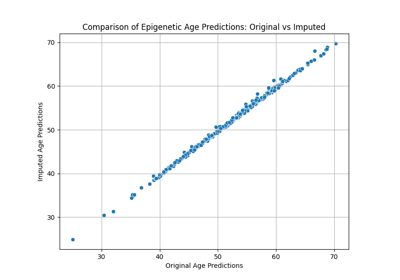
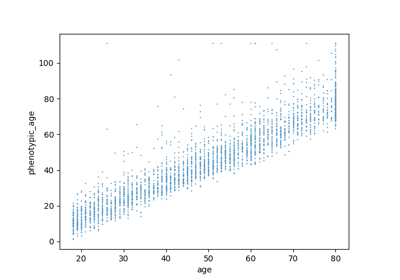
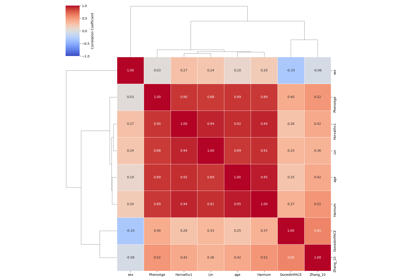
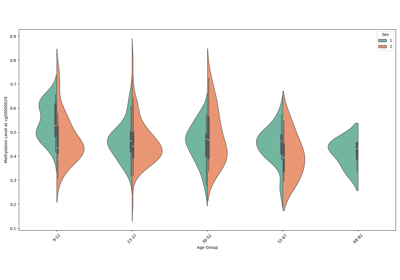
This document showcases functions in visualize.py for visualizing quality control results, DNA methylation, and aging clock/model predictions