Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Exploring the Challenge Data¶
This example shows you how to load and explore the challenge data with biolearn
Loading up the data for the competition¶
from biolearn.data_library import DataLibrary
challenge_data = DataLibrary().get("BoAChallengeData").load()
The challenge data has methylation data¶
import pandas as pd
pd.options.display.max_columns = 6
challenge_data.dnam
The challenge data also has proteomic data¶
You can learn more about what the protein identifies in our reference¶
from biolearn.util import get_data_file
reference = pd.read_csv(get_data_file("reference/alamar_reference.csv"))
reference
Some of the data overlaps while some does not but all the metadata is combined¶
You can easily run several models on them¶
from biolearn.mortality import run_predictions
prediction_dict = {
"Horvathv1": "Predicted",
"Hannum": "Predicted"
}
predictions = run_predictions(challenge_data, prediction_dict)
predictions
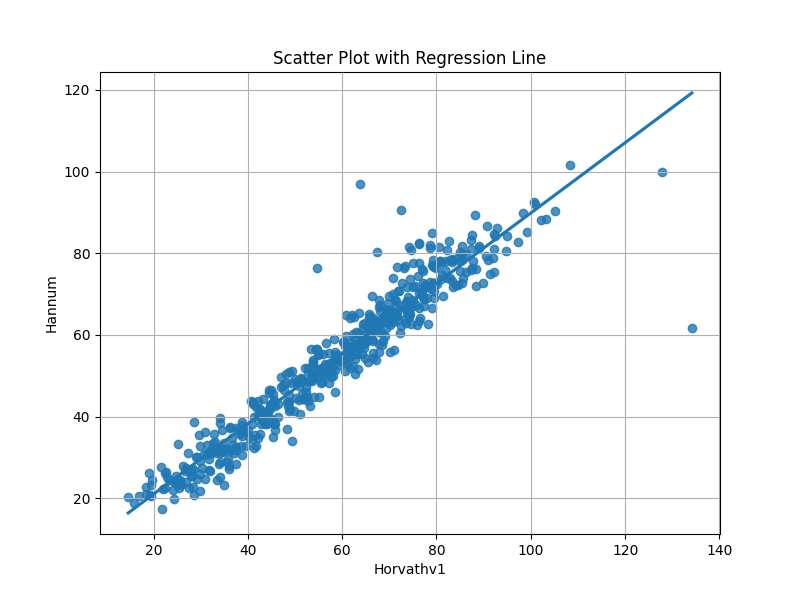
We can then compare the output from the two models¶
import matplotlib.pyplot as plt
import seaborn as sns
# Assuming your dataframe is named 'df'
# df should have columns 'Horvathv1' and 'Hannum'
# Create a scatter plot with a regression line
plt.figure(figsize=(8, 6))
sns.regplot(x='Horvathv1', y='Hannum', data=predictions, ci=None)
plt.title('Scatter Plot with Regression Line')
plt.xlabel('Horvathv1')
plt.ylabel('Hannum')
plt.grid(True)
plt.show()
Total running time of the script: (1 minutes 14.006 seconds)
Estimated memory usage: 13937 MB