Note
Go to the end to download the full example code. or to run this example in your browser via Binder
“Transcriptomic Clock” using GEO Data¶
This example loads transcriptomic data from GEO, calculates the predicted age using a transcriptomic clock, and plots it against chronological age.
First load up some transcriptomic data from GEO using the data library¶
from biolearn.data_library import DataLibrary
data = DataLibrary().get("GSE134080").load()
Now run the transcriptomic clock to predict age¶
from biolearn.model_gallery import ModelGallery
rna_model = ModelGallery().get("TranscriptomicPredictionModel", imputation_method="none")
predicted = rna_model.predict(data)
Plot the results against the chronological age using seaborn¶
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import pearsonr
predicted_values = predicted['Predicted']
true_values = data.metadata['age']
correlation, _ = pearsonr(true_values, predicted_values)
print(f"Pearson Correlation Coefficient: {correlation}")
sns.set_theme(style="whitegrid")
sns.regplot(x=true_values, y=predicted_values, scatter_kws={'alpha':0.5})
plt.title('Transcriptomic Prediction Model')
plt.xlabel('Chronological Age')
plt.ylabel('Transcriptomic Age')
plt.show()
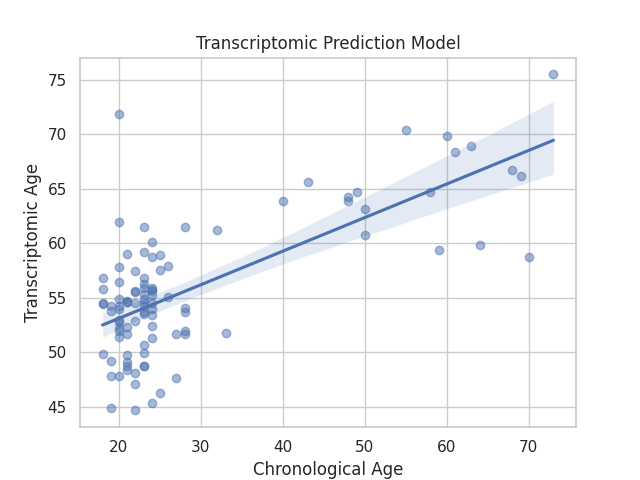
Pearson Correlation Coefficient: 0.7025211774998293
Total running time of the script: (0 minutes 13.164 seconds)
Estimated memory usage: 691 MB