Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Clock/model visualizations using GEO datasets¶
This example demonstrates the built-in aging clock/model function to visualize different plots on clock(s)/model(s) predictions.
Import required classes and functions¶
from biolearn.data_library import DataLibrary
from biolearn.model_gallery import ModelGallery
from biolearn.visualize import (
plot_clock_correlation_matrix,
plot_clock_deviation_heatmap,
plot_age_prediction,
plot_health_outcome
)
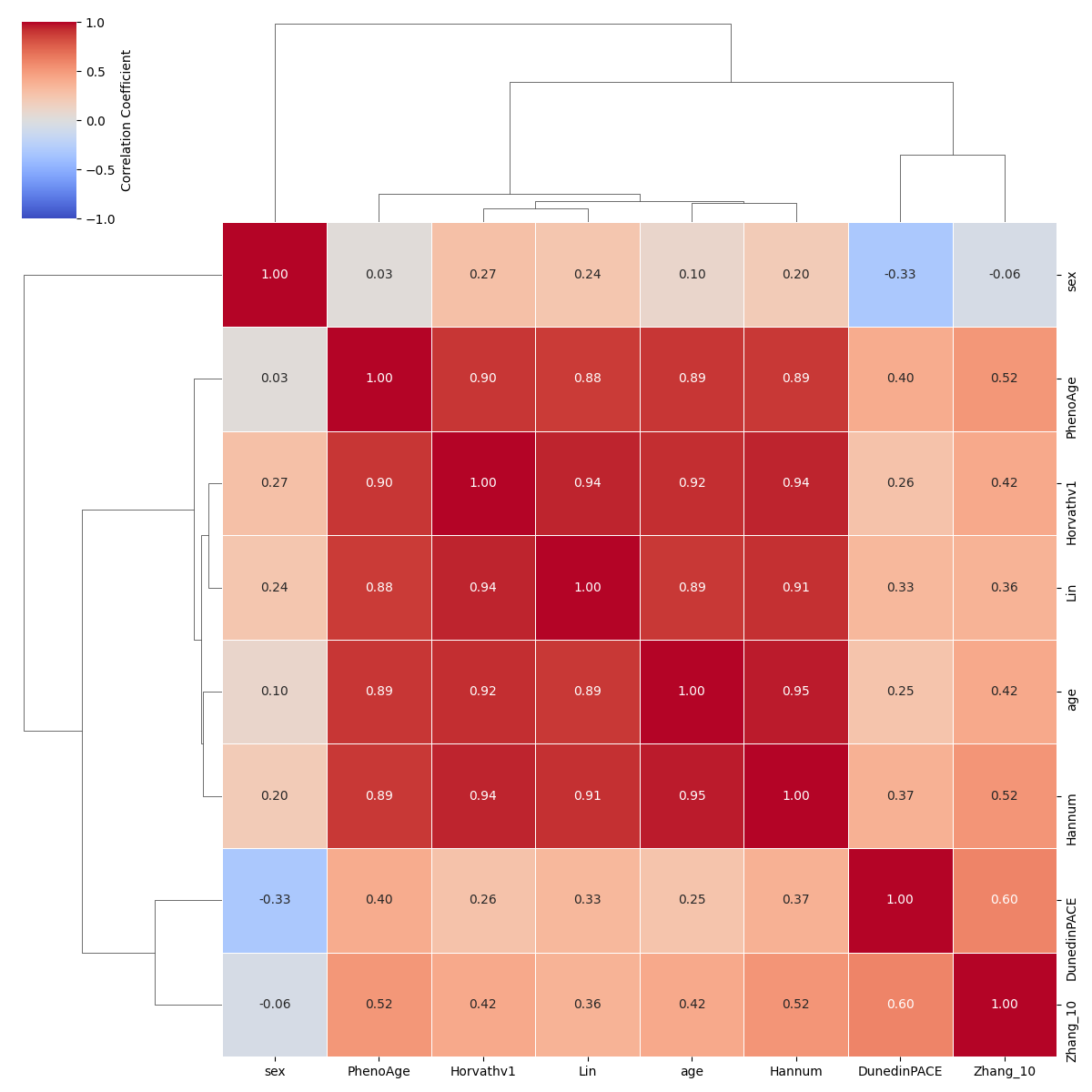
Visualize a correlation matrix across aging clocks/models¶
# Load an appropriate GEO dataset for the models
data = DataLibrary().get("GSE120307").load()
# Create a list of ModelGallery objects to be analyzed.
modelnames = ["Horvathv1", "Hannum", "PhenoAge", "DunedinPACE", "Lin", "Zhang_10"]
models = [ModelGallery().get(names) for names in modelnames]
plot_clock_correlation_matrix(
models=models,
data=data,
)
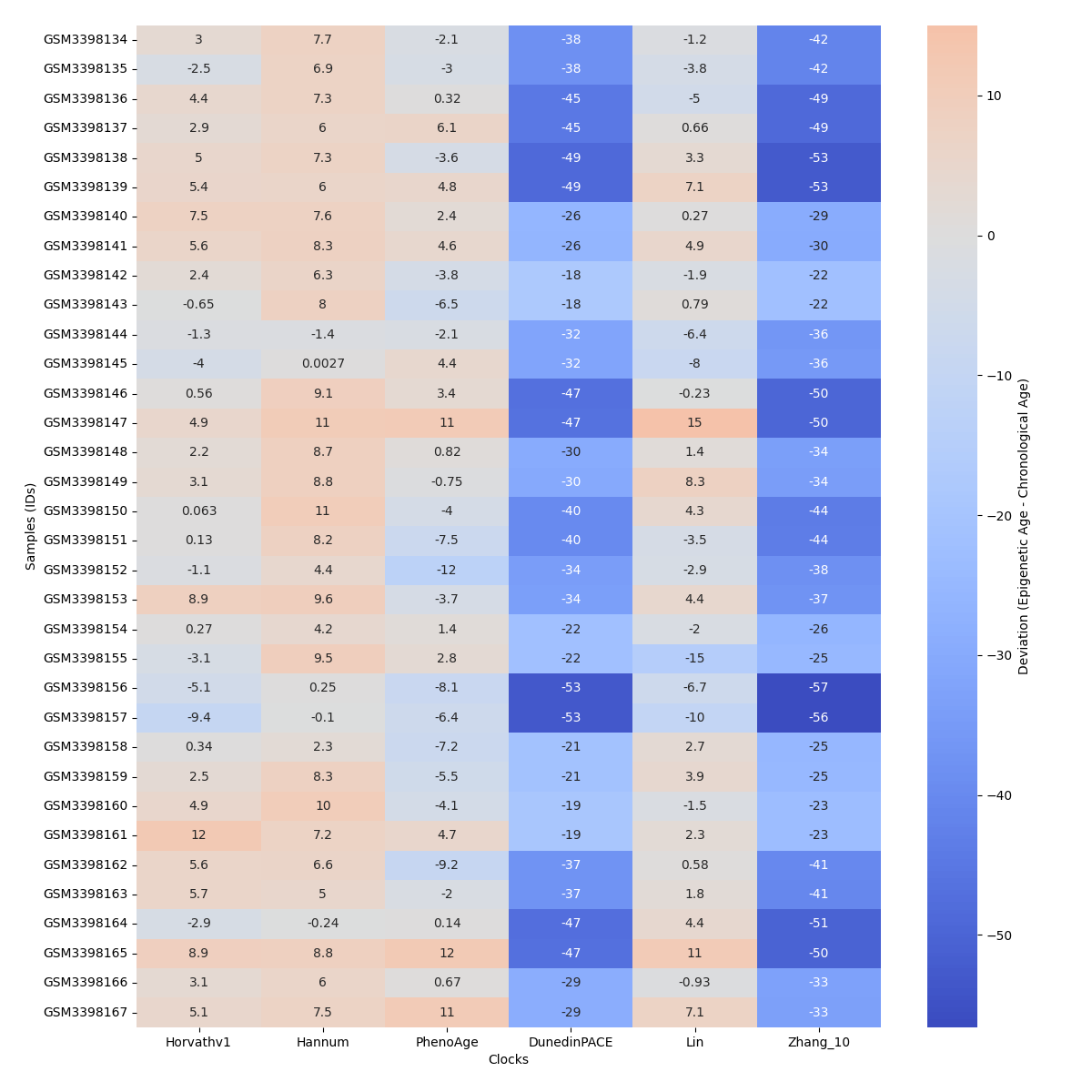
Visualize clock/model chronological age deviations across samples in a heatmap¶
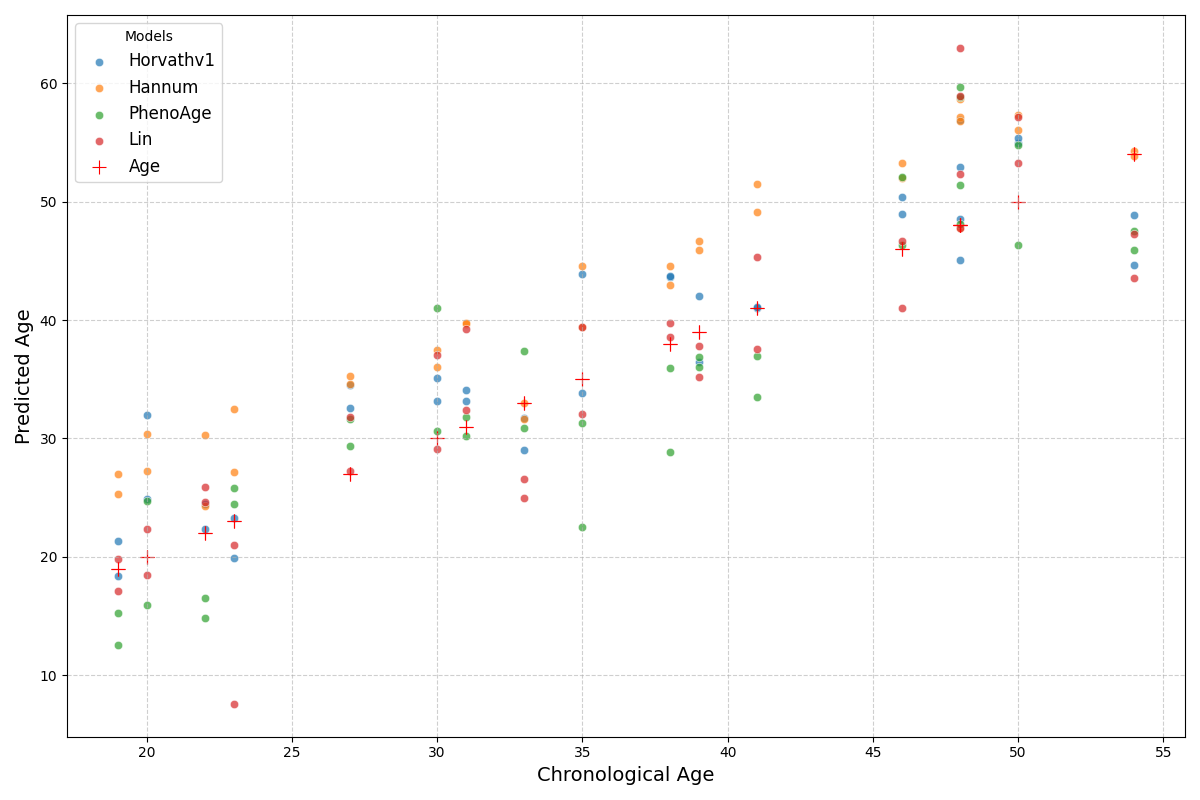
Visualize aging clock/model predictions against chronological age¶
# Use appropriate clocks/models
modelnames = ["Horvathv1", "Hannum", "PhenoAge", "Lin"]
age_prediction_models = [ModelGallery().get(name) for name in modelnames]
plot_age_prediction(
models=age_prediction_models,
data=data,
)
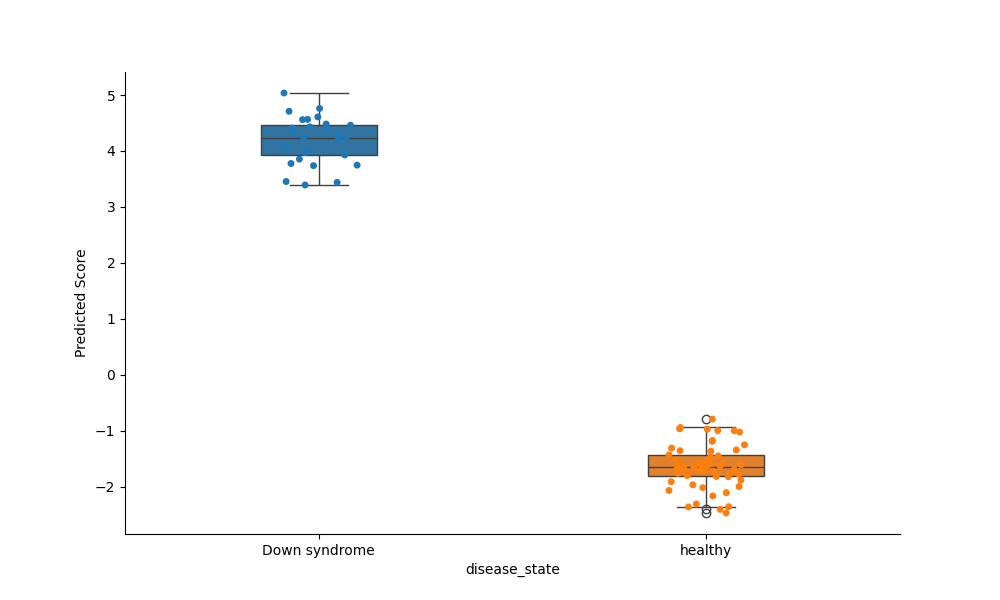
Visualize model predictions against its corresponding health outcome¶
# Load an appropriate GEO dataset for the corresponding model
down_syndrome_data = DataLibrary().get("GSE52588").load()
model = [ModelGallery().get("DownSyndrome")]
plot_health_outcome(
models=model,
data=down_syndrome_data,
# Provide the health outcome column name
health_outcome_col="disease_state",
)
Total running time of the script: (0 minutes 38.914 seconds)
Estimated memory usage: 1077 MB