Note
Go to the end to download the full example code. or to run this example in your browser via Binder
“Epigenetic Clocks” in GEO Data¶
This example loads a DNA Methylation data from GEO, calculates multiple epigenetic clocks, and plots them against chronological age.
First load up some methylation data from GEO using the data library¶
from biolearn.data_library import DataLibrary
#Load up GSE41169 blood DNAm data
data_source = DataLibrary().get("GSE41169")
data=data_source.load()
Now run three different clocks on the dataset to produce epigenetic clock ages¶
from biolearn.model_gallery import ModelGallery
gallery = ModelGallery()
#Note that by default clocks will impute missing data.
#To change this behavior set the imputation= parameter when getting the clock
horvath_results = gallery.get("Horvathv1").predict(data)
hannum_results = gallery.get("Hannum").predict(data)
phenoage_results = gallery.get("PhenoAge").predict(data)
Finally extract the age data from the metadata from GEO and plot the results using seaborn¶
import seaborn as sn
import pandas as pd
actual_age = data.metadata['age']
plot_data = pd.DataFrame({
'Horvath': horvath_results['Predicted'],
'Hannum': hannum_results['Predicted'],
'PhenoAge': phenoage_results['Predicted'],
"Age": actual_age
})
plot_data.index=plot_data['Age']
sn.relplot(data=plot_data, kind="scatter");
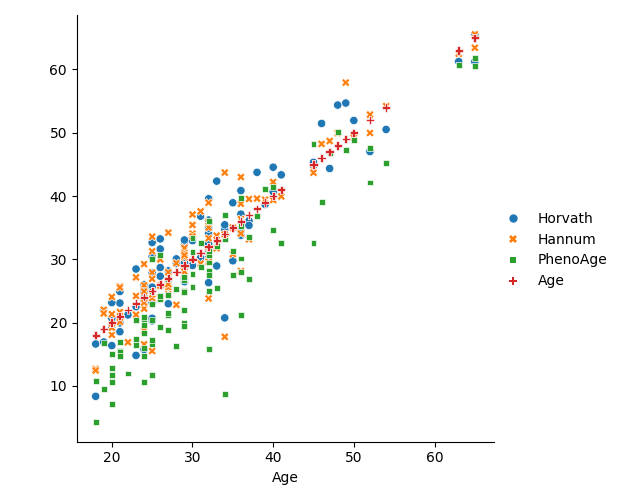
<seaborn.axisgrid.FacetGrid object at 0x1483f05c0>
Total running time of the script: (0 minutes 14.214 seconds)
Estimated memory usage: 1896 MB