Biolearn¶
Biolearn enables easy and versatile analyses of biomarkers of aging data. It provides tools to easily load data from publicly available sources like the Gene Expression Omnibus, National Health and Nutrition Examination Survey, and the Framingham Heart Study. Biolearn also contains reference implementations for common aging clocks such as the Horvath clock, DunedinPACE, and many others that can easily be run in only a few lines of code. You can read more about it in our paper.
Biolearn is developed and supported by several organizations and individuals, especially Biomarkers of Aging Consortium, Methuselah Foundation, and VOLO Foundation.
We are hosting a 2024-2025 Challenge series on the Synapse platform, where participants will be asked to predict chronological age, mortality, and multi-morbidity, with total awards of $200k+. Learn more at Synapse!.
If you use Biolearn in your research, please cite our preprint:
Ying, K., Paulson, S., Perez-Guevara, M., Emamifar, M., Martí nez, M. C., Kwon, D., Poganik, J. R., Moqri, M., & Gladyshev, V. N. (2023). Biolearn, an open-source library for biomarkers of aging. bioRxiv. https://doi.org/10.1101/2023.12.02.569722
Get started with Biolearn
See where Biolearn is headed and how to contribute
Join the Biomarkers of Aging Challenge!
Featured examples¶
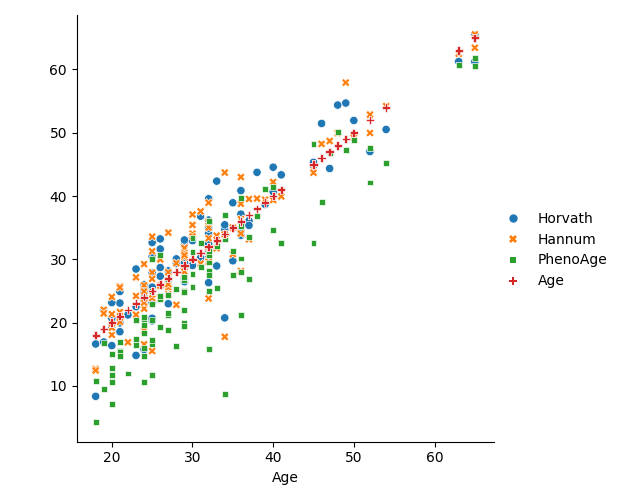
Demonstrate computation of several epigenetic clocks
Show how the clocks compare with chronological age
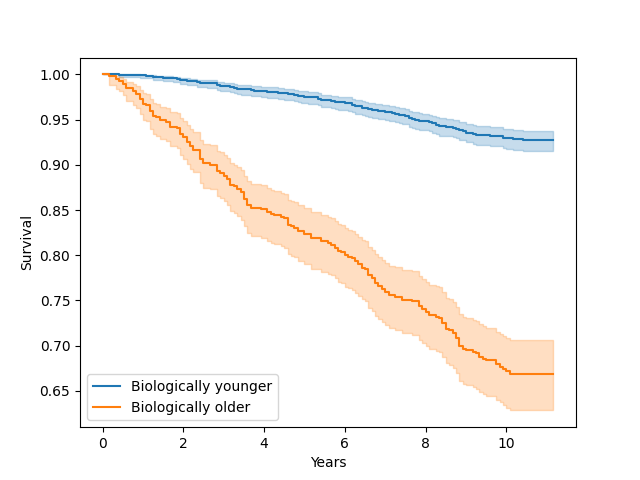
NHANES survival plotting with biological age
Using NHANES data shows relationship between various data points and survival