Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Performing custom imputations¶
This example demonstrates the ease of custom imputation using the biolearn library.
Load methylation data from a GEO dataset to use as a reference for imputation¶
from biolearn.data_library import DataLibrary, GeoData
# Load the reference dataset and compute averages for imputation
reference_dataset = DataLibrary().get("GSE40279").load()
reference_averages = reference_dataset.dnam.mean(axis=1)
Load up a target dataset and run the imputation¶
# Load a second dataset for imputation
target_dataset = DataLibrary().get("GSE51057").load()
# Perform imputation using the reference averages
from biolearn.imputation import impute_from_standard
imputed_data = impute_from_standard(target_dataset.dnam, reference_averages)
imputed_dataset = GeoData(target_dataset.metadata, imputed_data)
Now run clock predictions on the dataset before and after¶
# Using a model from the gallery, compare the epigenetic age before and after imputation
from biolearn.model_gallery import ModelGallery
clock_model = ModelGallery().get("Horvathv1")
original_age_predictions = clock_model.predict(target_dataset)
imputed_age_predictions = clock_model.predict(imputed_dataset)
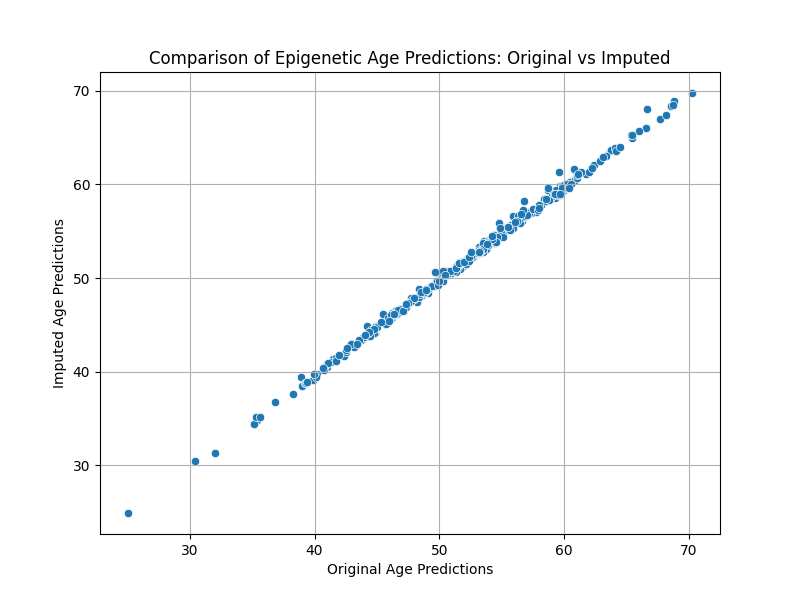
Visualize the comparison of age predictions¶
import seaborn as sns
import pandas as pd
import matplotlib.pyplot as plt
# Prepare data for visualization
comparison_data = pd.DataFrame({
'Original': original_age_predictions['Predicted'],
'Imputed': imputed_age_predictions['Predicted']
})
# Create a scatter plot to compare the results
plt.figure(figsize=(8, 6))
sns.scatterplot(x='Original', y='Imputed', data=comparison_data)
plt.title("Comparison of Epigenetic Age Predictions: Original vs Imputed")
plt.xlabel("Original Age Predictions")
plt.ylabel("Imputed Age Predictions")
plt.grid(True)
plt.show()
You can also build an imputation decorator to bundle with the clock¶
from biolearn.model import ImputationDecorator
# Define a custom imputation method using reference averages
def custom_impute_method(dnam_data, needed_cpgs):
return impute_from_standard(dnam_data, reference_averages, needed_cpgs)
# Wrap the clock model with the imputation decorator
decorated_clock = ImputationDecorator(clock_model, custom_impute_method)
You get the same results as running the function directly on your dataset¶
# Predict epigenetic age using the decorated clock (with imputation)
decorated_clock_predictions = decorated_clock.predict(target_dataset)
# Verify that the results are the same as the direct imputation approach
same_results = all(decorated_clock_predictions == imputed_age_predictions)
print(same_results)
True
Total running time of the script: (1 minutes 6.382 seconds)
Estimated memory usage: 8890 MB