Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Proteomic Organ Age Clock¶
This example loads proteomic data and runs it through the OrganAge clock
First load the COVID-19 serum proteomics data using the data library¶
from biolearn.data_library import DataLibrary
data_source = DataLibrary().get("Covid19_Filbin_Plasma")
data = data_source.load()
Now run the organ age clocks on the proteomic data¶
from biolearn.model_gallery import ModelGallery
gallery = ModelGallery()
chronological_results = gallery.get("OrganAge1500Chronological").predict(data)
Prepare data for visualization¶
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
plot_data_dict = {'age_range': data.metadata['age_range']}
ORGANS = ['Organismal', 'Multi-organ', 'Brain', 'Heart', 'Immune']
for organ in ORGANS:
plot_data_dict[organ] = chronological_results[organ]
plot_data = pd.DataFrame(plot_data_dict)
plot_data = plot_data.dropna(subset=['age_range'])
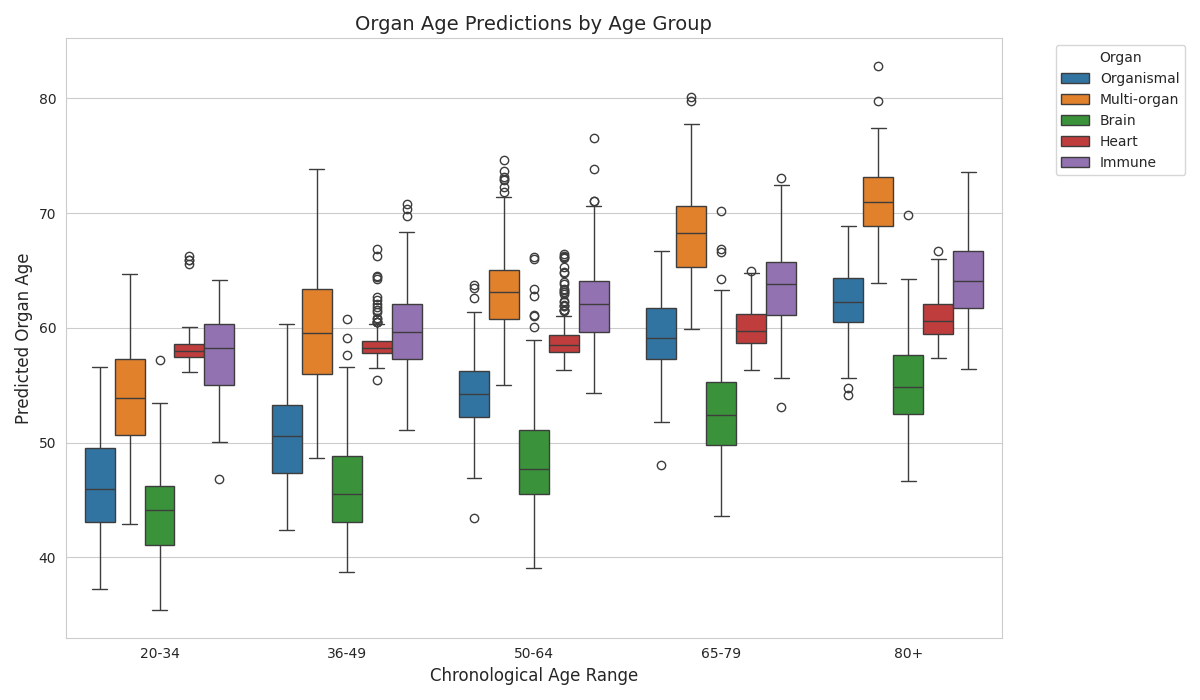
Create boxplot visualization¶
sns.set_style("whitegrid")
fig, ax = plt.subplots(figsize=(12, 7))
# Sort age ranges properly
age_order = sorted(plot_data['age_range'].unique())
plot_data_long = plot_data.melt(
id_vars=['age_range'],
value_vars=ORGANS,
var_name='Organ',
value_name='Predicted_Age'
)
sns.boxplot(
data=plot_data_long,
x='age_range',
y='Predicted_Age',
hue='Organ',
order=age_order,
ax=ax
)
ax.set_xlabel('Chronological Age Range', fontsize=12)
ax.set_ylabel('Predicted Organ Age', fontsize=12)
ax.set_title('Organ Age Predictions by Age Group', fontsize=14)
ax.legend(title='Organ', bbox_to_anchor=(1.05, 1), loc='upper left')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 3.364 seconds)
Estimated memory usage: 184 MB